Watch a TEDx Talk by Dr. Adam Marcus about targeting metastasis.
The development of targeted therapy represents an exciting new approach to cancer treatment. A small number of these drugs have already been FDA approved and several others are currently in clinical trials.
In August of 2017, researchers from the University of Colorado Cancer Center performed a study on lung cancer patients. Researchers looked at both the overall survival and the progression-free survival, also known as the time it take for a tumor to restart its growth, of the patient. The researchers found that, "With immunotherapies, there were better outcomes for deeper responses, but it didn’t break down the same way as with targeted treatment against ALK-positive cancer." This means, that for their study with lung cancer patients, the data suggested that the deeper the response, the longer it will last. This suggests that the depth of the response may allow physicians and patients to better compare treatment effectiveness. 1
Topics covered in the following sections:
- Targeted Therapies vs Conventional Chemotherapy
- Kinase Inhibitors: Kinases are enzymes that add phosphate groups onto proteins. Because they control many cellular processes, abnormal activity can result in the development of cancer.
- Antibody Treatments: Antibodies are naturally occuring proteins of the immune system. 'Designer' antibodies are used to treat a variety of cancers.
- Angiogenesis Inhibitors: Angiogenesis, the development of blood vessels, is critical to the growth of almost all types of cancer.
- Proteasome Inhibitors: Proteasomes are responsible for the destruction of proteins. Inhibition of proteasome activity be used to treat some types of cancer.
- Molecular Receptor Blockers: Messages are sent between and within cells. Abnormal messaging can lead to cancer and some treatments are designed to interfere with the transmission of these types of messages.
- Histone Acetyl Transferases (HATs) and Histone Deacetylases (HDACs): DNA serves as a blueprint for the activities in cells. Access to this information is controlled by modifications of the DNA, including addition/removal of small chemical groups (acetyl groups) to the proteins associated with DNA.
- Histone Deacetlylase (HDAC) Inhibitors: The addition and removal of acetyl groups to histones leads to the creation of pattern of modifications that is critical to the activity of DNA. Manipulation of this 'histone code' can be used to treat cancer.
- Antisense Drugs: Antisense RNA is able to interfere with the activity of the RNA messages (mRNA) produced when DNA is transcribed.
- Ribozymes: Ribozymes are RNA molecules with enzymatic (catalytic) activity. This is in contrast with the much better understood protein-based enzymes.
- Enzyme Activators: Cancer can result in the inactivation of critical enzymes. Some cancer treatments function by turning these inactive enzymes back on.
- Know the Flow: Targeted Therapy
Watch the interview with Dr. Alex Adjei.
Perhaps the most promising theme in the development of more specific cancer drugs is the targeting of cancer-specific processes, instead of processes common to all cells. Because these drugs are not directly toxic, and because they only affect cancer cells, they offer the hope of being highly specific with few side effects.2 The specificity of current drugs does have one drawback. Blocking a single pathway in a cancer cell may be enough to slow it down, but it often does not inhibit the cancer enough to kill it. Therefore, many specific cancer drugs are currently being used together with traditional chemotherapy. The combination of a highly specific cancer drug that is able to attack a tumor's weaknesses, and standard chemotherapy to deliver a powerful attack on the tumor, may prove to be an excellent means of treating cancer. This section will offer some examples of drugs that have been/are being developed to target cancer-specific processes and spare normal cells.
Targeted Therapies vs Conventional Chemotherapy
Dr. Fadlo Khuri discusses the difference between chemotherapy and targeted therapy. Watch the full interview with Dr. Fadlo Khuri.
Cancer cells and normal cells have much in common in terms of the internal machinery that allows them to carryout the many activities necessary to stay alive. Chemotherapy drugs effectively target processes that cancer cells need to grow and divide, such as the ability of the cancer cells to replicate their DNA. However, many normal cells, like the cells that line the digestive tract, also need to replicate. In short, though chemotherapy drugs are particularly toxic to cancer cells, they also damage healthy cells. The use of standard chemotherapy therefore produces many, and often severe, side effects. Furthermore, these side effects sometimes prevent patients from being able to take high enough doses to fight the cancer most effectively. While chemotherapy drugs are quite effective treatments for many forms of cancer, researchers have been working diligently to produce drugs that target the processes of cancer cells specifically so as to leave healthy cells unharmed. The accumulation of knowledge about the specific differences between normal and cancerous cells has allowed for the development of treatments targeted at cancer-specific activities.
One of the most fundamental changes found in cancer cells is the presence of mutations in the genes that are responsible for causing cell growth (oncogenes). The defective proteins produced by these altered genes are prime candidates for targeted therapy. As an example, some cancers are caused in part by mutant proteins that send constant signals into the cell causing cell division. Drugs that block only the mutant form of the protein but do not interfere with the activity of the normal version would only affect cancer cells, and would leave healthy cells untouched. Alternatively, many cancers result when genes that normally prevent cell growth (tumor suppressors) are inactivated or turned off. Drugs that "fix" the activity of these proteins would repair the damaged cancer cells, but theoretically have no effect on normal cells.
Kinase Inhibitors
Functions of Kinases
The kinases include a large number of enzymes that regulate the activity of other proteins and, more indirectly, the activities of cells. All kinases add phosphate groups to other molecules, often other proteins, in the cell. Protein phosphorylation, the addition of a phosphate group to a side chain of an amino acid3, is an important regulatory action. Because each phosphate group carries two negative charges, the addition of one of a phosphate can cause a change in the shape of a protein. Altered protein shape is often correlated with altered activity of the protein. The ability to change the conformation of a protein between two different shapes allows for regulated control of that protein's activity. Phosphorylation (addition of phosphate groups to a protein) by kinases is a reversible process,and proteins can be dephosphorylated (removal of phosphate group) by enzymes called protein phosphatases.3These two groups of enzymes often work together to 'turn on' and 'turn off' pathways within cells. The kinases play an important role in many intracellular signaling pathways, including those that control cell growth and cell division.
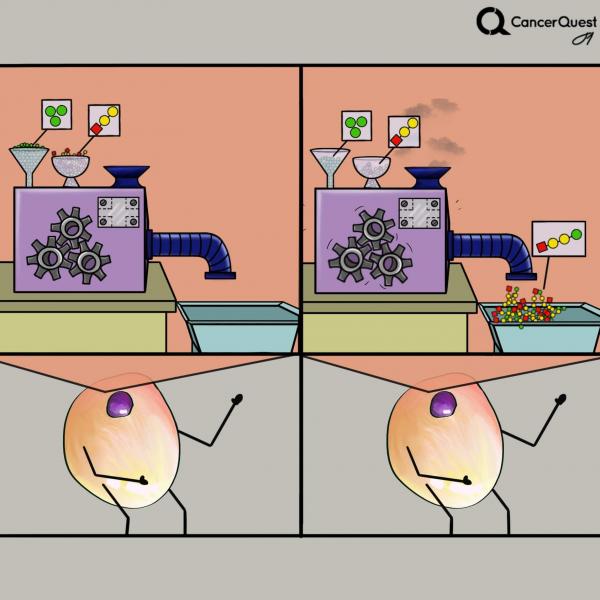
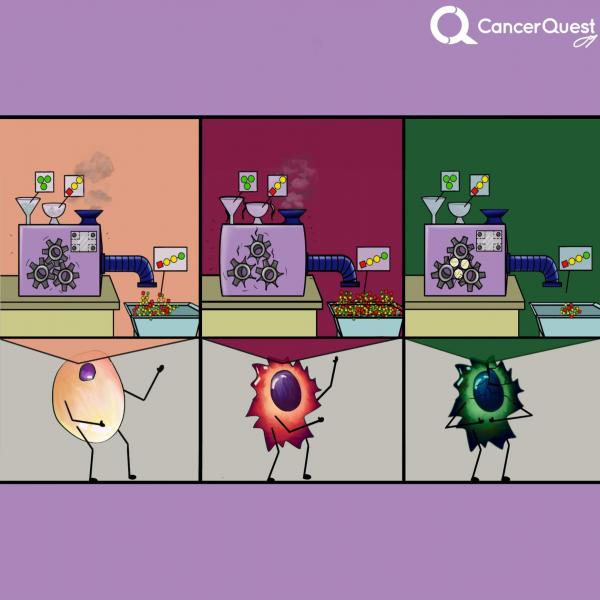
Kinases are enzymes (molecular machines) that add phosphates from ATP (green) to targets. There are many different kinds of kinases.
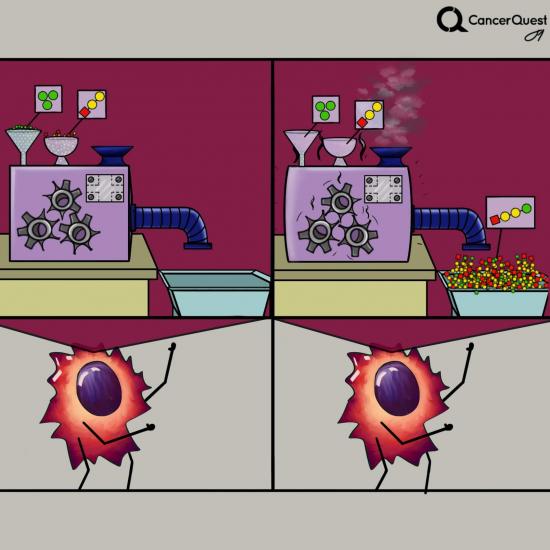
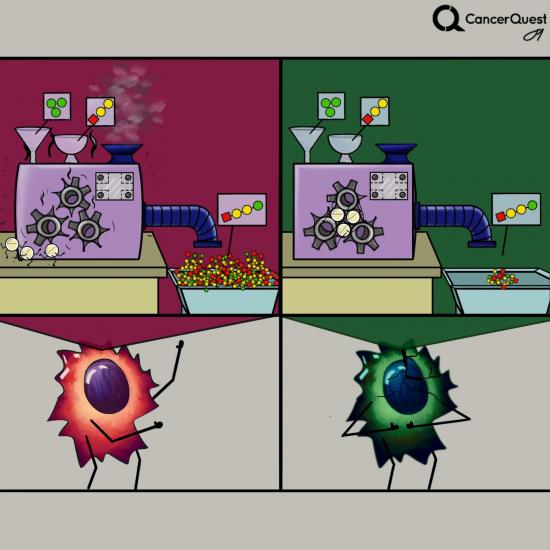
In cancer, one or more kinases are often damaged (mutated) and get stuck in the active form. The constant activity can lead to increased cancer cell growth and survival.
Learn more about kinases by watching the full interview with Dr. William Hahn.
Kinases and Cancer
Cell growth and cell cycle pathways are constitutively activated in cancer cells. The normal controls exerted by the kinase/phosphatase enzymes no longer function.One key feature of cancer cells is their ability to reproduce in the absence of external signals such as growth factors. In the normal process, growth factors that are excreted by other cells bind to receptors on the cell surface, stimulating the cell to divide. Cancerous cells turn on the pathway in the absence of the growth factor. This may occur because of a mutation in a kinase or phosphatase gene. In one example, chronic myeloid leukemia, a particular chromosomal translocation (termed the Philadelphia chromosome) has been identified that creates a novel kinase that is 'on' all the time. The pathway that this kinase controls is, in effect, stuck in the 'on' position. This leads to proliferation of the cancerous cells.
Several new cancer treatments are designed to inhibit aberrantly activated kinases within cancer cells in an effort to prevent cell division. The following drugs are kinase inhibitors used to treat cancer.
- Sorafenib (Nexavar®)
- Imatinib (Gleevec®, Glivec®)
- Gefitinib (Iressa®)
- Lapatinib (Tykerb®)
- Dasatinib (Sprycel®)
- Nilotinib (Tasigna®)
- Temsirolimus (Toricel®)
- Erlotinib (Tarceva®)
Antibody-based Treatments
Antibodies are proteins produced by immune cells called B-lymphocytes (B-cells). Antibodies bind to certain molecules on foreign objects (called antigens) like pollen or bacteria that enter the body. This tells the body that the object is foreign and marks it for destruction and removal. The target molecules are often but not always proteins. Antibodies may also bind to antigens located on the surface of other cells, including cancer cells. 4
The interaction between antibody and antigen is very precise and is just like the interaction between a lock and key. Any given B-cell can produce only one type of antibody but our cells contain millions of different B-cells. Because of this, there are many different kinds of antibodies in our bodies. In fact, our bodies have the ability to respond to each of the many thousands or even millions of different possible antigens to which we may be exposed during our lifetime. When a particular B-cell encounters the appropriate antigen it reproduces to create a large number of cells. Some of the resulting cells are very long-lived. They can live for years after the initial 'challenge' with antigen and provide the immune system with a form of memory. A second exposure to the same antigen leads to to a much more rapid and vigorous response. This long term memory is the basis of vaccinations. The rest of the cells are short-lived cells that produce large amounts of antibodies.5, 4
As shown below, antibodies are composed of four proteins, two larger chains and two smaller chains, which are represented in purple. They are shaped like Ys and each side is identical so that an antibody is able to bind to two copies of its target, one per each short arm of the Y.
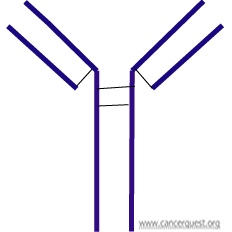
Any given antigen can lead to the activation of many different B-cells and the production of many different antibodies, all directed against different portions of the same target. A mixture of different antibodies is said to be polyclonal.
In the laboratory, it is possible to produce and purify large amounts of a single type of antibody, produced by a single type of B-cell. These antibodies are said to be monoclonal. The antibody based treatments developed so far utilize monoclonal antibodies.5, 4
Details of Antibody Structure
All antibodies (also called Immunoglobulins - written as Ig) share some common features. Each contains two smaller proteins (light chains) and two larger proteins (heavy chains). The four proteins that make up the antibody are held together by disulfide (S-S) bonds between cysteine amino acids. Each of the four chains of the antibody contains a constant region and a variable region. The constant (Fc) region contains domains that allow other cells of the immune system to recognize that antibody. The small red pieces represent the light chains and the blue pieces represent the heavy chains. On the left arm of the antibody the variable regions are colored in green (there is also a variable region on the other arm in the same location - not colored here). The disulfide (S-S) bonds that hold the molecule together are shown.
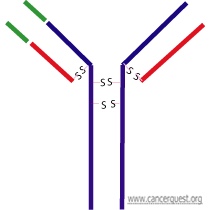
Types of antibodies
There are five major kinds of antibody, called isotypes. The isotype of antibody is determined by its constant region. The five types of antibody are IgG, IgD, IgA, IgE, and IgM; each isotype is associated with a distinct immune response. For example, antibodies of the IgG isotype often recruit cytotoxic (killer) T-cells, that destroy target cells. The antibody shown in the figure above is of the IgG1 isotype.
The variable regions of antibodies recognize and bind to specific antigens. It is possible for antibodies produced by more than one type of B-cell to recognize a particular target. For example, a bacterial invader would have many target proteins on its surface and would be recognized by many different antibodies within the body. Even a single protein (or other type of antigen) may be recognized by many different antibodies.
Antibody responses
Polyclonal antibodies arise when the body is presented with an antigen and many different B-cells are activated to produce antibodies. The result is a mixture of antibodies that would all recognize different parts of the target antigen.
Monoclonal antibodies are those that are derived from a single type of B cell. Monoclonal antibodies are used as anti-cancer drugs.
Antibodies in Cancer Treatment
Our immune system performs its job remarkably well. It is believed that many abnormal cells are eliminated by the immune system before they can progress to cancer. Unfortunately many cancer cells tend to go unnoticed by the immune system because they originate from normal body cells. Despite the fact that they behave like foreign organisms within our bodies, cancer cells often do not elicit a significant immune response. The use of the immune system and its products in the treatment of cancer has yielded promising results.
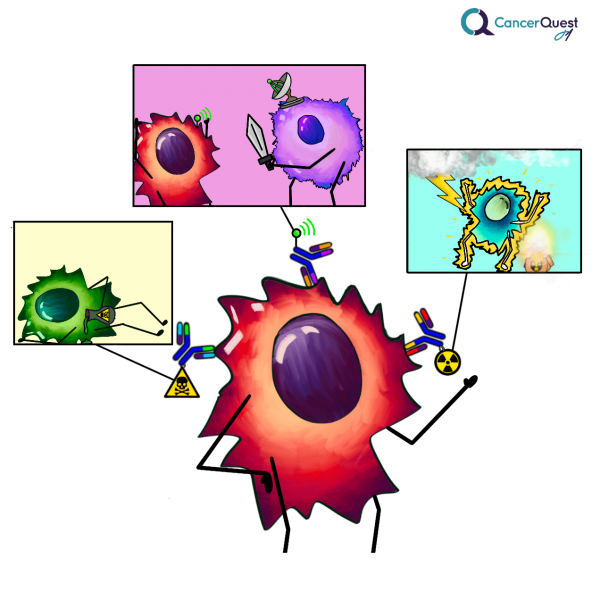
Antibodies can be used to inhibit the growth of cancer cells in several different ways:
-
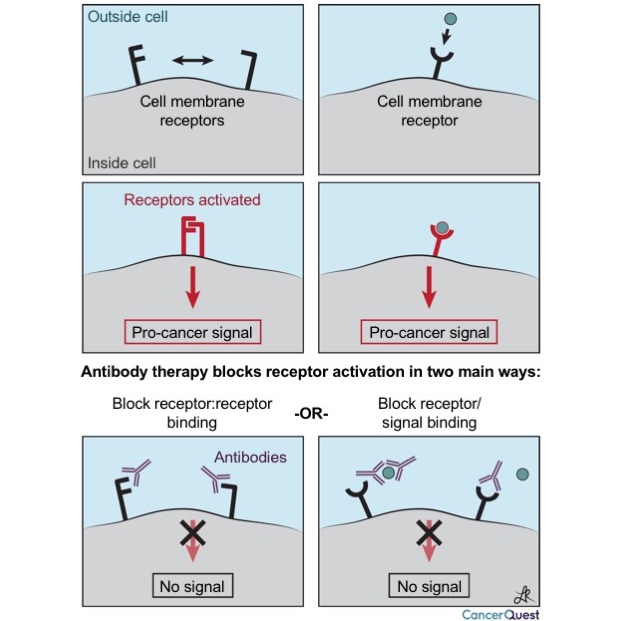
Blockage of receptors: These antibodies may function as a blockade to the receipt of required growth signals.
-
Antibodies that stimulate cell killing: These antibodies function by targeting proteins on the surface of cancer cells. The antibodies themselves mark the cell for destruction by cells of the immune system. This process is termed antibody dependent cellular cytotoxicity (ADCC).4
-
Immunotoxins: This approach utilizes antibodies to target toxic molecules to the cancer cells. These toxic molecules can be proteins that inhibit cellular activities or radioactive compounds that cause DNA damage and the induction of apoptosis.
The first type of action is shown below. Antibodies are very specific in what they target, and can block interactions between proteins and signaling molecules. Two different types of cancer driving signals are shown below, the first is caused by the interaction of two proteins on the surface of a cancer cell, and the second is triggered when a signal from outside a cell binds to a receptor (like an antenna) on the surface of the cancer cell.
The graphic below show more ways that antibodies can be used to help kill cancer cells.
Antibody-Based Cancer Drugs
There are many antibody treatments being used to treat different types of cancer. Antibodies that are currently approved for cancer treatment include:
Bexxar® (tositumomab)
Campath® (alemtuzumab)
Herceptin® (trastuzumab)
Rituxan® (rituximab)
Zevalin® (ibritumomab)
Bexxar® (tositumomab)
Erbitux® (cetuximab)
Vectibix® (panitumumab)
See Angiogenesis Inhibitors for information on another antibody treatment - Avastin®.
Table of Antibody Treatments An easy to use table of antibody treatments; including trade name, generic name, and type. Links to more information are provided.
A Closer Look at Herceptin®
Many tumors overproduce proteins necessary for cell growth and division. An example is the overexpression of the HER2 receptor protein, which is found in excessive quantities in some breast and ovarian cancer cases. More information on HER-2/neu. In cancers that do produce too much of this receptor, Herceptin® may help to prevent the growth of primary and metastatic tumors.
Herceptin® is a monoclonal antibody specifically engineered to bind to the HER2 protein. It is not known exactly how Herceptin® functions to stop the growth of sensitive tumors, but there is evidence that it may have more than one mechanism of action.6 It is thought that Herceptin® may prevent tumor growth by inhibiting the binding of extracellular growth signals to receptors on the cell surface. The normal signaling process and activity of Herceptin® are shown in the animation below.
Herceptin® may induce antibody dependent cellular cytotoxicity (ADCC). Other mechanisms are also possible.7, 8
Angiogenesis Inhibitors
Like all cells, cancer cells require a constant supply of nutrients and oxygen in order to grow and divide. Without an adequate blood supply tumors will not grow. Tumors produce factors that stimulate the formation of blood vessels to provide them with the food and oxygen they need.
The process of blood vessel formation is termed angiogenesis. One way to inhibit the development of blood vessels is to block the signals coming from the tumor. Cancer cells either directly produce signals that cause blood vessels to form or they 'trick' nearby, non-tumor, cells to send out the signals.
One possible benefit of this type of cancer treatment is that they should have less negative effects (side effects) on the patient. Angiogenesis occurs at high levels during fetal development, the menstrual cycle and in wound healing. The treatments might be expected to interfere with these processes but should not harm most normal dividing cells.
Unlike conventional chemotherapy, the cancer cells are not directly killed by the treatment. The targets of several of these treatments are normal processes controlled by normal cells (such as the cells that form blood vessels), not the tumor cells themselves. Cancer cells are very unstable but the targets of these drugs should be much less likely to change in the presence of the drugs. This might make the cancer cells less likely to become resistant to these types of treamtent.
There are other approaches to the inhibition of angiogenesis which are actively being investigated:
- Matrix Metalloproteinases Inhibitors: The degradation of the extracellular matrix that surrounds all cells is an important process in the in the formation of new blood vessels. As described in detail in the section on metastasis, this process is also integral to the spread of tumor cells to distant locations in the body.Growing blood vessel cells secrete enzymes called matrix metalloproteinases (MMPs) that are able to digest the extracellular matrix and allow blood vessels to invade the area and supply the tumor with nutrients. Inhibition of this process is the target of several drugs.
- Endothelial Cell Inhibitors: Several different drugs are under investigation that inhibit angiogenesis by acting to prevent the growth or activities of the endothelial cells that form the blood vessels.
- Inhibitors of Angiogenesis Activation: The drugs in this class of angiogenesis inhibitors work by blocking the cascade of events that cause blood vessels to form.
The targeted drugs currently being developed may actually affect more than one target. Sometimes this is intentional and other times it is an unavoidable side-effect of the drug.
Some FDA approved drugs that work (at least in part) by preventing angiogenesis:
Proteasome Inhibitors
Proteasomes are a subcellular organelle found throughout the cytosol, nucleus, endoplasmic reticulum (ER), and lysosomes of eukaryotic cells.9 They are cylindrical in shape and are comprised of several different proteolytic (protein digesting) enzymes. The functions of proteasomes include:
- Digestion of unassembled proteins.
- Digestion of damaged or misfolded proteins.
- The generation of peptides that are recognized by the immune system.
- Regulation of the cellular lifespan (half-life)of regulatory proteins such as those that control the cell cycle.10
Over 80% of a cell's proteins are processed by proteasomes.11
Proteasomes act on proteins that have been specifically tagged for degradation by attachment to another protein called ubiquitin. Degradation signals are generally hidden in a properly folded protein, but become accessible when the protein is misfolded or denatured. When these signals are exposed, enzymes add another small protein (ubiquitin; shown as a yellow sphere below) to the target. The ubiquitinated protein is then recognized by a receptor protein on the proteasome and the target protein is taken into the proteasome and digested. The fragmented protein is then released from the proteasome as shown below.10
Proteasomes control the half-life of many short-lived regulatory proteins, such as those involved in the cell cycle. Therefore, proteasome malfunction can lead to abnormal regulation of the cell cycle and uncontrolled cell proliferation. The cell cycle is controlled by both positive and negative signals. An alteration in the proteins involved in these controls can tip the balance and may play a major role in the development and progression of cancer. In a normal cell, proteasomes break down proteins that inhibit the cell cycle, such as cyclin-dependent kinase inhibitors (CKI). 9
Inhibition of proteasome function causes cell cycle arrest and cell death. Tumor cells are more susceptible to these effects than normal cells, in part because they divide more rapidly and in part because many of their normal regulatory pathways are disrupted. The mechanism for the differential response of normal and cancer cells to proteasome inhibition is not fully understood. Overall, cancer cells are more susceptible to proteasome inhibitors and, as a result, these inhibitors may be an effective treatment for certain cancers.9
The first proteasome inhibitor to enter clinical trials as a cancer treatment was PS-341 (now: MLN-341). The drug is produced by Millenium Pharmaceuticals. The drug specifically interacts with a key amino acid (a threonine) within the active site (catalytic site) of the proteasome. Although this interaction blocks only 1 of the 3 catalytic activities of the proteasome, the resulting inhibition seems to be sufficient to prevent the degradation of ubiquitinated proteins.12 On May 13, 2003 PS-341, now known as bortezomib or Velcade™ became the first proteasome inhibitor to be approved for use in the US. It is now used to treat several blood cell cancers.13
A second proteasome inhibitor was isolated from a marine bacteria. The agent is called salinosporin or salinosporamide and it is currently being studied. 14
Learn more about borezomib (VELCADE®)
A Closer Look at Proteosome Structure and Function
The proteasome has a structure that is related to its function. Each proteasome consists of a cylindrical central 20S core composed of four stacked rings containing seven proteins each. The outer two rings are identical and composed of 'alpha' subunits, and the middle two are identical and contain 'beta' subunits. The beta subunits exhibit active protein digesting activity. The digestion occurs on the sides of the enzymes facing the inner chamber.
The core of the proteasome is bound on each end to a (19S) regulatory protein. The 19S cap proteins are responsible for the recognition of proteins to be degraded. In a process that requires energy (ATP), tagged proteins are unfolded and fed into the inner chamber. The proteins are then digested into peptides 6 to 9 amino acids in length and released. The peptides can then be recycled. Together the 19S caps and the 20S core make up the human proteasome.
Proteasomes and the immune system.
Some digested protein fragments enter the endoplasmic reticulum (ER) where they are joined to proteins that function in generating immune responses. The complex of the peptides and the immune system protein (MHC protein) is then transported to the surface of the cell. This complex can act as a flag for cells of the immune system and lead to an immune response. An example of this would be the processing of proteins produced by viruses within an infected cell.
Molecular Receptor Blockers
Our cells constantly monitor their surroundings for the presence (or sometimes an absence) of regulatory molecules in the environment. These signals control decisions regarding cell division, movement and even death. One way in which the external environment is monitored is through receptor molecules located in the membrane of the cell. These proteins act like cellular antennas. Upon binding of a signal molecule, the receptor molecules transmit information to other molecules within the cytoplasm and nucleus resulting in an appropriate response. An example is the HER-2/neu oncogene product, that functions as a growth factor receptor. This gene is discussed more in the section on oncogenes.15 Many different diseases, including cancer, can be traced back to dysfunctional signaling pathways. In cancer these malfunctions may lead to unregulated cell division and the development of tumors. Particular signaling pathways are often affected in a given type of cancer. Drugs designed to inhibit these specific signaling pathways promise to inhibit cancer growth without harming normal cells. Blockage of the receptors is an active area of clinical investigation. 15
Specific receptor inhibition remains a relatively new but promising area of medical research. One potential problem with the receptor inhibitors are the long-term side effects of these drugs. The treatments are relatively new so the side effects are also not well described at this time. As a general rule the side effects documented to date are much less severe than conventional chemotherapy treatments.
There are now several drugs that work by binding to cell surface proteins and receptors. Additional specific receptor antagonists target hormone receptors located within cells. Information on these inhibitors is located in the Hormonal section of the Cancer Treatments section.
Breast Cancer - Selective Estrogen Receptor Modifiers (SERM's)
Prostate Cancer - Selective Androgen Receptor Modifiers (SARM's)
Drugs that block blood vessel growth are described in the section on angiogenesis inhibitors.
Histone Acetyl Transferases (HATs) and Histone Deacetylases (HDACs)
With few exceptions, such as red blood cells, all of our cells contain two full copies of our genome. Each chromosome is composed of a single piece of double-stranded DNA containing millions or even hundreds of millions of nucleotide pairs. These molecules are very thin and extremely long. Each cell contains approximately 2 meters (~6 feet) of DNA. All of this DNA must be packaged in such a way that it fits into the cell nucleus, a very small space. To accomplish this feat, the negatively charged DNA is condensed and tightly wrapped around positively charged proteins called histones.16 Eight histone proteins (two copies each of four different proteins) form a core around which the DNA winds. The histone proteins, along with the DNA wrapped around it, form a structure called a nucleosome. Each nulceosome holds only about 150 base pairs of DNA so there are thousands of these structures along the length of each chromosome. As shown in the figure below, the nucleosomes are connected together by small stretches of DNA and look like beads on a string. Nucleosomes can further condense in the presense of additional, non-histone proteins to form chromatin.17
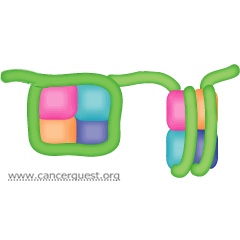
The ability of a particular transcription factor to bind to its target gene is, in part, dependent on modifications that are made to the histone proteins. Enzymes called histone acetyl transferases (HATs) alter the chromatin (DNA:protein) structure by adding short carbon chains to some histone proteins. This alteration changes the structure of the DNA:histone interaction loosening up the 'bead', allowing transcription factors to bind. A separate group of enzymes, histone deacetlyases (HDACs), is responsible for removing the acetyl groups from the histones, shutting down transcription.
When the histones are deacetylated, the positive charges are restored and the DNA binds more tightly to the nucleosome. Deacetylation leads to a repression of transcription because the necessary transcription factors, regulatory factors, and RNA polymerase complex are unable to gain access to the DNA. 18
Abnormal activity of HDACs has been observed in several different types of cancer, such as acute promyelocytic leukemia, acute myelogenous leukemia, non-Hodgkin lymphoma, and some types of colorectal and gastric carcinomas. When these enzymes act incorrectly, they can prevent the transcription of key genes. This process appears to be an important step in the tumorigenic process in some forms of cancer.19, 20
Histone Deacetlylase (HDAC) Inhibitors
HDAC inhibitors have been shown to alter the growth of several different forms of cancer.20 These molecules, many of which have been isolated from natural sources, have demonstrated the ability to inhibit proliferation, induce differentiation, and cause apoptosis in tumor cells.
One exciting feature of these molecules is that they have demonstrated little toxicity in preliminary tests. Although much of the testing for these potential therapies has been performed only in cell culture and animal models, the results of these tests have been very promising and several clinical trials are underway as a result. HDAC inhibitors have been shown to affect many different types of solid and hematological cancers in research studies, including neuroblastoma, melanoma, leukemia (including APL and myeloid), breast cancer, prostate cancer, lung cancer, ovarian cancer, and colon cancer.20
HDAC inhibitors are grouped into categories based on their chemical structures. The exact mechanism through which these compounds work is still poorly understood, but is a focus of current research. It is thought, though, that each inhibitor acts to inhibit a specific HDAC; this in turn has a specific effect upon gene expression, cell cycle regulation, proliferation, differentiation, and apoptosis. Scientists also believe that HDAC inhibitors cause changes in the cytoskeleton, which further aids in their anti-tumor activity. Many of these compounds are effective in very low doses and appear to act upon specific regions of the genome, altering the transcription of only select sets of genes. The true efficacy of HDAC inhibitors is yet to be determined, in testing they have shown the potential to be effective, selective treatments for some forms of cancer. These compounds may be used in combination with other anti-cancer agents.20
The FDA recently approved a new HDAC inhibitor in 2006 called Vorinostat (Zolinza®)
Antisense Drugs
As discussed in detail in the section on Cell Function, our genes consist of long strings of nucleotides along the DNA in our chromosomes. When there is a requirement for any particular gene to be expressed, the process of transcription leads to the production of an mRNA message from that gene.
Unlike the double-stranded structure of DNA, messenger RNA molecules are single-stranded. Like DNA molecules, these single-stranded RNA molecules are capable of binding to complementary nucleic acid strands. Therefore, if another single stranded string of nucleic acid bases with a sequence complementary to the sequence of a given mRNA (the antisense sequence) enters the cell, it will bind to that specific mRNA and inactivate it. The bound mRNA is not suitable for translation and is degraded.
Antisense oligonucleotide drugs are one of the newer areas of drug development. This technology offers researchers the ability to target almost any cellular process with complete specificity. If a protein is helping a cancer cell to grow, then the appropriate antisense oligonucleotide could be used to prevent that protein from ever being made. Because antisense oligonucleotides are so specific, it is unlikely that any other protein in the body would be affected. This specificity should result in a reduction of the side effects often seen with conventional cancer treatments.21
The function of antisense RNA is shown in the animation below. The first part of the animation depicts the normal translation process of the mRNA (purple) and the second part shows the binding of an antisense RNA (red) to the mRNA (purple) leading to a blockage of protein production.
The full mechanism of this inactivation is not entirely understood, but it may be related to the fact that double stranded RNA occurs only rarely in normal cells. Since the instructions to make each protein are carried by a unique mRNA, these messages can be individually inactivated or "knocked out" by these complementary sequences.21
No antisense cancer drugs have been approved by the FDA but many clinical trials are underway testing a agents directed against different targets and in different cancer types.22, 23, 24
View a list of clinical trials involving antisense drugs (NCI).
Ribozymes
Enzymes are organic catalysts that speed up biological reactions. Although the majority of known enzymes are proteins, some RNA molecules, ribozymes, also display catalytic activity. Ribozymes are involved in a variety of cellular processes, but their most interesting property from the standpoint of cancer therapy is their ability to cleave messenger RNA (mRNA) molecules. When an mRNA is cleaved, it can no longer be translated to produce protein. By targeting the mRNAs encoding proteins with pathological roles in cancer, ribozymes can slow or inhibit cancerous growth.
Ribozymes possess two key regions that participate in the cleavage of mRNA. One region binds to the target RNA sequence and the other is responsible for cleavage of the target site. The binding of the ribozyme to the target involves pairing of the two RNA molecules and is very specific. This allows for the design of ribozymes directed against particular mRNA molecules. If the nucleotide sequence of an mRNA is known, a ribozyme can be designed to specifically cleave that mRNA. The results of the human genome project promise to be critical in identifying specific genes/proteins important in cancer, giving ribozyme designers many targets. By targeting particular mRNAs, ribozymes can combat specific stages of cancerous growth, including cell proliferation, drug resistance, and metastasis.
The animation below shows the binding of a ribozyme to mRNA and its cleavage of the mRNA into two pieces.
Some of the genes that have been targeted include:
- ras -an oncogene that drives cancer cell proliferation25, 26 More on ras
- c-fos - encodes a protein that promotes transcription of other genes 18
- MDR1 - encodes a transmembrane protein involved in resistance to chemotherapy drugs.27, 28 More on MDR
- MMP-9 - an enzyme involved in tumor metastasis 16
- VEGFR-1 - a receptor for the vascular endothelial derived growth factor. A ribozyme treatment, Angiozyme® (RPI.4610), was the first ribozyme drug developed.29 It was tested in kidney cancer but is no longer in clinical testing.
No ribozyme drugs have been approved by the FDA.
Check for clinical trials of ribozyme-based treatments (NCI).
A Closer Look at Ribozymes
ANGIOZYME was the first synthesized ribozyme to be studied in human trials. It was designed to combat the process of angiogenesis by cleaving the mRNA that produces the VEGF1 (Vascular Endothelial Growth Factor) receptor. Reduction in the amounts of this receptor should lead to the inhibition of blood vessel formation in the tumor and therefore inhibit tumor growth.
ANGIOZYME was developed by Sirna Therapeutics (now closed) in collaboration with Chiron (purchased in 2006 by Novartis). It was tested in clinical trials for several cancers, including breast and colon cancers. There were limited significant toxicities related to drug use in trials and the occurrence of side effects was low. 30Unfortunately, the clinical responses to Angiozyme were not very good. It is not currently being evaluated in clinical trials.
Enzyme Activators
Enzymes are proteins that work to speed up chemical reactions in the body. Enyzmes can work to build up or break down molecules needed for cell growth and maintenance. Shown below is an animation showing an enzyme that joins together two biomolecules to make one larger one.
Many chemotherapy agents function by inhibiting key enzymzes. Examples of agents that inhibit enzymes include the topoisomerase inhibitors Irinotecan and Topotecan and many others.
The activation or supplementation of enzymes that normally function to limit cell growth can also decrease tumor cell division. An example of this type of drug is listed below:
- Asparaginase (Elspar®)
Know the Flow: Targeted Therapy
Know the Flow is an educational game for you to test your knowledge. To play:
- Drag the appropriate choices from the column on the right and place them in order in the boxes on the left. Note that you will only use five of the six choices to complete the game.
- When done, click on 'Check' to see how many you got correct.
- For incorrect answers, click on 'Description' to review information about the processes.
- To try again, choose 'Reset' and start over.
Please visit us on a larger screen to play this game.
- 1 C. E. McCoach G. M. Blumenthal L. Zhang A. Myers S. Tang R. Sridhara P. Keegan R. Pazdur R. C. Doebele D. Kazandjian. Exploratory analysis of the association of depth of response and survival in patients with metastatic non-small cell lung cancer treated with a targeted therapy or immunotherapy. 2017 Aug 2. [ANNALS OF ONCOLOGY]
- 2 Gibbs J. "Mechanism-based target identification and drug discovery in cancer research." Science (2000) 287: 1969-1973 [PUBMED]
- 3ab Becker, Hardin, Kleinsmith. The World of the Cell. San Francisco: Benjamin Cummings, 2003.
- 4abcd Goldsby R, Kindt T, Osborne B. Kuby Immunology. 4th ed. WH Freeman & Company, New York: 2000.
- 5ab Microbiology and Immunology Online. Produced by the University of South Carolina School of Medicine. Accessed August 26, 2010 [http://pathmicro.med.sc.edu/book/immunol-sta.htm]
- 6 Genentech Product Information [http://www.gene.com]
- 7 Albanell J, Arribas J, Baselga J, Codony-Servat J, Molina MA, Rojo F. "Trastuzumab (Herpceptin), a humanized anti-HER2 receptor monoclonal antibody, inhibits basal and activated HER2 ectodomain cleavage in breast cancer cells." Cancer Research (June 2001). 61: 4744-4749. [PUBMED]
- 8 Barnes M. "Response of metastatic breast cancer to trastuzamab." Lancet (2000). 355: 160-161. [PUBMED]
- 9abc Adams J. "Development of the proteosome inhibitor PS-341." The Oncologist (2002). 7: 9-16. [PUBMED]
- 10ab Schwartz AL, Ciechanover A. "The ubiquitin-proteasome pathway and pathogenesis of human diseases." Annu Rev Med (1999). 50:57-74. [PUBMED]
- 11 Geradts J, Ingram CD. "Abnormal expression of cell cycle regulatory proteins in ductal and lobular carcinomas of the breast." Modern Pathology (2000). 13(9): 945-953. [PUBMED]
- 12 Adams J. "Adams J. Proteasome inhibition: a novel approach to cancer therapy. Trends Mol Med. 2002;8(4 Suppl):S49–S54. doi:10.1016/s1471-4914(02)02315-8[PubMed]
- 13 Einsele H. Bortezomib Recent Results Cancer Res. 2010;184:173-87. [PUBMED]
- 14 Chauhan D, Catley L, Li G, et al. (2005). "A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib". Cancer Cell 8 (5): 407-419 [PUBMED]
- 15ab Downward, J. "The ins and outs of signaling" Nature (June 14, 2001). 411: 759-762. [PUBMED]
- 16ab Hua J and Muschel RJ. "Inhibition of matrix metalloproteinase 9 expression by a ribozyme blocks metastasis in a rat sarcoma model system." Cancer Res (1996). 56: 5279-82. [PUBMED]
- 17 Ribozyme Pharmaceuticals [http://www.rpi.com]
- 18ab Scanlon KJ, et al. "Ribozyme-mediated cleavage of c-fos mRNA reduces gene expression of DNA synthesis enzymes and metallothionein." PNAS (1991). 88 (23): 10591-5. [PUBMED]
- 19 Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. "Histone deacetylases and cancer: causes and therapies." Nat Rev Cancer (2001). 1(3): 194-202. [PUBMED]
- 20abcd Richon VM, O'Brien JP. "Histone deacetylase inhibitors: a new class of potential therapeutic agents for cancer treatment." Clin Cancer Res (2002). 8(3): 662-4. [PUBMED]
- 21ab Isis Pharmaceuticals [http://www.isip.com]
- 22 Caruso G, Caffo M, Raudino G, Alafaci C, Salpietro FM, Tomasello F. Antisense oligonucleotides as innovative therapeutic strategy in the treatment of high-grade gliomas. Recent Pat CNS Drug Discov. 2010 Jan;5(1):53-69. [PUBMED]
- 23 Cassell A, Grandis JR. Investigational EGFR-targeted therapy in head and neck squamous cell carcinoma. Expert Opin Investig Drugs. 2010 Jun;19(6):709-22. [PUBMED]
- 24 Patel MP, Masood A, Patel PS, Chanan-Khan AA. Targeting the Bcl-2. Curr Opin Oncol. 2009 Nov;21(6):516-23. [PUBMED]
- 25 Ohta Y, et al. "H-ras ribozyme-mediated alteration of the human melanoma phenotype." Ann NY Acad Sci (1994). 716: 242-53. [PUBMED]
- 26 Wang CH, Tsai LJ, Tsao YP, Hsieh JT, Chien WW, Liao CL, Wang HW, Liu HS, Chen SL. "Recombinant adenovirus encoding H-ras ribozyme induces apoptosis in laryngeal cancer cells through caspase- and mitochondria-dependent pathways." Biochem Biophys Res Commun. 2002 Nov 15;298(5):805-14. [PUBMED]
- 27 Scanlon KJ, et al. "Ribozyme-mediated reversal of the multidrug-resistant phenotype." PNAS (1994). 91(23): 11123-7. [PUBMED]
- 28 Nagata J, Kijima H, Hatanaka H, Asai S, Miyachi H, Abe Y, Yamazaki H, Nakamura M, Watanabe N, Mine T, Kondo T, Scanlon KJ, Ueyama Y. "Reversal of drug resistance using hammerhead ribozymes against multidrug resistance-associated protein and multidrug resistance 1 gene." Int J Oncol. 2002 Nov;21(5):1021-6. [PUBMED]
- 29 Weng DE, Usman N. Angiozyme: a novel angiogenesis inhibitor. Curr Oncol Rep. 2001 Mar;3(2):141-6. [PUBMED]
- 30 Weng DE, Usman N. "Angiozyme: a novel angiogenesis inhibitor." Curr Oncol Rep. 2001 Mar;3(2):141-6. [PUBMED]